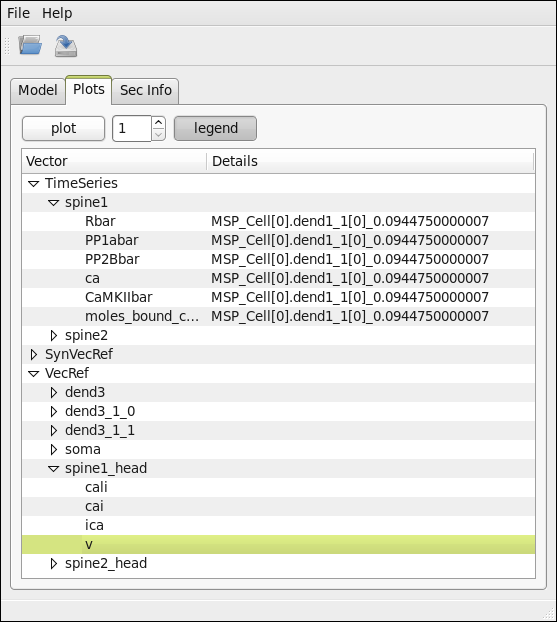
Saving and loading simulations’ reults¶
HDF structure¶
Neuronvisio stores simulation’s results using the hdf standard, using PyTables This is very handy when you simulation takes a long time to run and you want to inspect again the results, without re-run it.
The file has a structure shown in the following _static
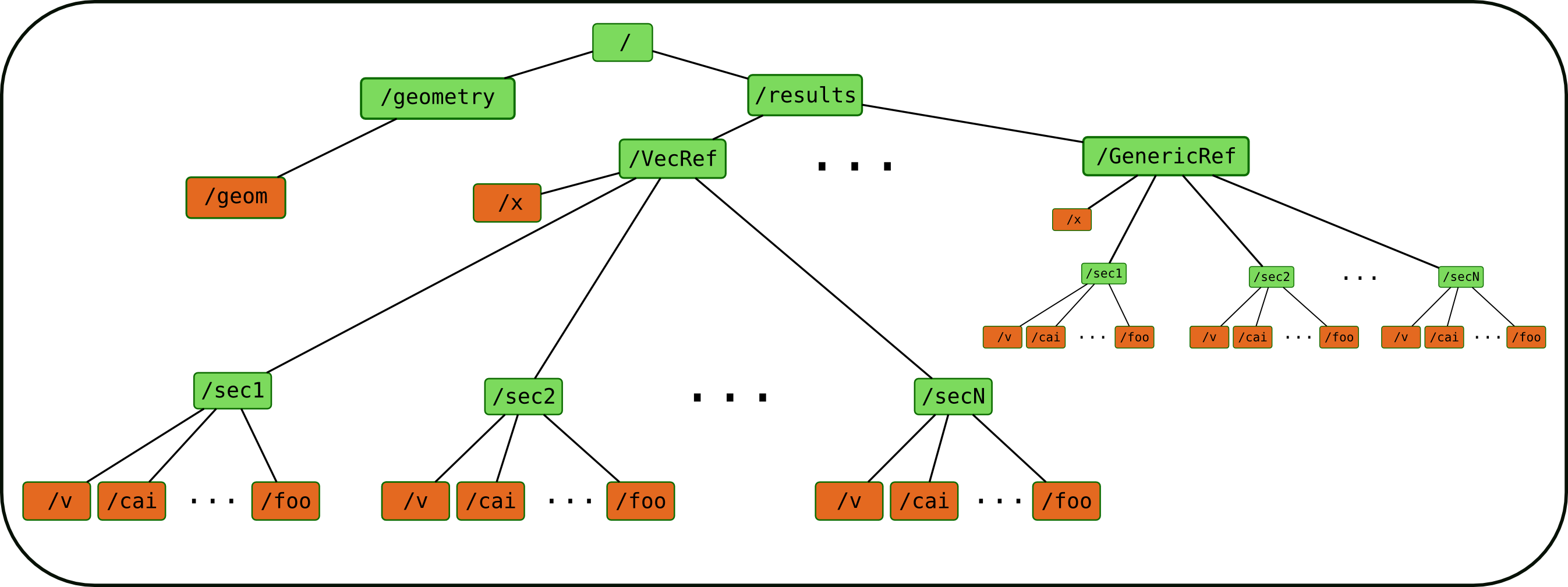
The Refs is the data structure used by Neuronvisio. The VecRef is the specialized one. It is possible to add more
Ref subclassing the manager.BaseRef.
Using the manager object to store the results of your simulation¶
This is a quick example how to save the simulation in neuronvisio:
# Model geometry already instantiated.
#
from neuronvisio.manager import Manager
manager = Manager()
manager.add_all_vecRef('v') # Adding vector for the variable v
# Perform your simulation
# ...
#
# file where to save the results
filename = 'storage.h5'
# Saving the vectors
manager.save_to_hdf(filename)
If you run a lot of simulations you want maybe to run the same script but without rewriting the same results. Manager has a nice method to help you called create_new_dir:
saving_dir = manager.create_new_dir() # Create a new dir per Simulation, ordered by Day.
hdf_name = 'storage.h5'
filename = os.path.join(saving_dir, hdf_name)
# Saving the vectors
manager.save_to_hdf(filename)
Loading a previous simulation¶
To load the results of a simulation you can start neuronvisio giving the path_to_the_hdf_file:
$ neuronvisio path/to/storage.h5
or you can just start neuronvisio and use the Load button:
$ neuronvisio
Saving your variables in storage.h5 and use Neuronvisio to plot them¶
The BaseRef can be used to store computational results which are not in NEURON vectors format. Every BaseRef object is contained in a group, which is specified by the group_id attribute. The group_id is than used by Neuronvisio to pair the saved vectors with the time vectors which is required to plot.
To subclass the BaseRef just create a class:
from neuronvisio.manager import BaseRef
class MyRef(BaseRef):
def __init__(self, sec_name=None, vecs=None, detail=None):
BaseRef.__init__(self)
self.group_id = 'MyGroup'
self.sec_name = sec_name
self.vecs = vecs
self.detail = detail
Then you can create it:
myRef = MyRef(sec_name=sec_name,
vecs=vecs,
detail=detail)
sec_name should be the name of the section where the variable is been recorded, vecs is a dictionary with the variable name as key and the python_list with the computed value. A numpy array or an HocVector is also accepted.
After that you can add to the manager using the manager.add_ref which takes two arguments:
- the myRef object
- the x variable.
The x variable is the independent one, usually the time, which will be used to plot the from the Neuronvisio graphical interface. If don’t need to supply your own time vector, because is the same of the main NEURON one, you can use the manager.groups[‘t’] which will return the NEURON time array:
manager.add_ref(myRef, x)
All together is:
class MyRef(BaseRef):
def __init__(self, sec_name=None, vecs=None, detail=None):
BaseRef.__init__(self)
self.sec_name = sec_name
self.vecs = vecs
self.detail = detail
myRef = MyRef(sec_name=sec_name,
vecs=vecs,
detail=detail)
manager.add_ref(myRef, x)
Then you just need to save the file where is more convenient for you:
filename = 'storage.h5'
# Saving the vectors
manager.save_to_hdf(filename)
When you reload the simulation you will have your variables back